Release 260111
This commit is contained in:
70
common/transformations/README.md
Normal file
70
common/transformations/README.md
Normal file
@@ -0,0 +1,70 @@
|
||||
|
||||
Reference Frames
|
||||
------
|
||||
Many reference frames are used throughout. This
|
||||
folder contains all helper functions needed to
|
||||
transform between them. Generally this is done
|
||||
by generating a rotation matrix and multiplying.
|
||||
|
||||
|
||||
| Name | [x, y, z] | Units | Notes |
|
||||
| :-------------: |:-------------:| :-----:| :----: |
|
||||
| Geodetic | [Latitude, Longitude, Altitude] | geodetic coordinates | Sometimes used as [lon, lat, alt], avoid this frame. |
|
||||
| ECEF | [x, y, z] | meters | We use **ITRF14 (IGS14)**, NOT NAD83. <br> This is the global Mesh3D frame. |
|
||||
| NED | [North, East, Down] | meters | Relative to earth's surface, useful for visualizing. |
|
||||
| Device | [Forward, Right, Down] | meters | This is the Mesh3D local frame. <br> Relative to camera, **not imu.** <br> 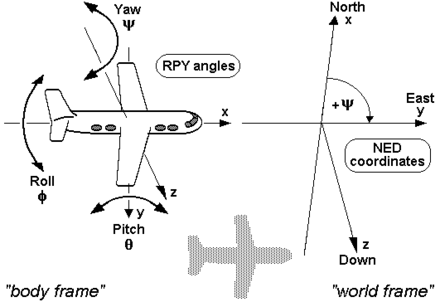|
|
||||
| Calibrated | [Forward, Right, Down] | meters | This is the frame the model outputs are in. <br> More details below. <br>|
|
||||
| Car | [Forward, Right, Down] | meters | This is useful for estimating position of points on the road. <br> More details below. <br>|
|
||||
| View | [Right, Down, Forward] | meters | Like device frame, but according to camera conventions. |
|
||||
| Camera | [u, v, focal] | pixels | Like view frame, but 2d on the camera image.|
|
||||
| Normalized Camera | [u / focal, v / focal, 1] | / | |
|
||||
| Model | [u, v, focal] | pixels | The sampled rectangle of the full camera frame the model uses. |
|
||||
| Normalized Model | [u / focal, v / focal, 1] | / | |
|
||||
|
||||
|
||||
|
||||
|
||||
Orientation Conventions
|
||||
------
|
||||
Quaternions, rotation matrices and euler angles are three
|
||||
equivalent representations of orientation and all three are
|
||||
used throughout the code base.
|
||||
|
||||
For euler angles the preferred convention is [roll, pitch, yaw]
|
||||
which corresponds to rotations around the [x, y, z] axes. All
|
||||
euler angles should always be in radians or radians/s unless
|
||||
for plotting or display purposes. For quaternions the hamilton
|
||||
notations is preferred which is [q<sub>w</sub>, q<sub>x</sub>, q<sub>y</sub>, q<sub>z</sub>]. All quaternions
|
||||
should always be normalized with a strictly positive q<sub>w</sub>. **These
|
||||
quaternions are a unique representation of orientation whereas euler angles
|
||||
or rotation matrices are not.**
|
||||
|
||||
To rotate from one frame into another with euler angles the
|
||||
convention is to rotate around roll, then pitch and then yaw,
|
||||
while rotating around the rotated axes, not the original axes.
|
||||
|
||||
|
||||
Car frame
|
||||
------
|
||||
Device frame is aligned with the road-facing camera used by openpilot. However, when controlling the vehicle it is helpful to think in a reference frame aligned with the vehicle. These two reference frames can be different.
|
||||
|
||||
The orientation of car frame is defined to be aligned with the car's direction of travel and the road plane when the vehicle is driving on a flat road and not turning. The origin of car frame is defined to be directly below device frame (in car frame), such that it is on the road plane. The position and orientation of this frame is not necessarily always aligned with the direction of travel or the road plane due to suspension movements and other effects.
|
||||
|
||||
|
||||
Calibrated frame
|
||||
------
|
||||
It is helpful for openpilot's driving model to take in images that look similar when mounted differently in different cars. To achieve this we "calibrate" the images by transforming it into calibrated frame. Calibrated frame is defined to be aligned with car frame in pitch and yaw, and aligned with device frame in roll. It also has the same origin as device frame.
|
||||
|
||||
|
||||
Example
|
||||
------
|
||||
To transform global Mesh3D positions and orientations (positions_ecef, quats_ecef) into the local frame described by the
|
||||
first position and orientation from Mesh3D one would do:
|
||||
```
|
||||
ecef_from_local = rot_from_quat(quats_ecef[0])
|
||||
local_from_ecef = ecef_from_local.T
|
||||
positions_local = np.einsum('ij,kj->ki', local_from_ecef, postions_ecef - positions_ecef[0])
|
||||
rotations_global = rot_from_quat(quats_ecef)
|
||||
rotations_local = np.einsum('ij,kjl->kil', local_from_ecef, rotations_global)
|
||||
eulers_local = euler_from_rot(rotations_local)
|
||||
```
|
||||
0
common/transformations/__init__.py
Normal file
0
common/transformations/__init__.py
Normal file
179
common/transformations/camera.py
Normal file
179
common/transformations/camera.py
Normal file
@@ -0,0 +1,179 @@
|
||||
import itertools
|
||||
import numpy as np
|
||||
from dataclasses import dataclass
|
||||
|
||||
import openpilot.common.transformations.orientation as orient
|
||||
|
||||
## -- hardcoded hardware params --
|
||||
@dataclass(frozen=True)
|
||||
class CameraConfig:
|
||||
width: int
|
||||
height: int
|
||||
focal_length: float
|
||||
|
||||
@property
|
||||
def size(self):
|
||||
return (self.width, self.height)
|
||||
|
||||
@property
|
||||
def intrinsics(self):
|
||||
# aka 'K' aka camera_frame_from_view_frame
|
||||
return np.array([
|
||||
[self.focal_length, 0.0, float(self.width)/2],
|
||||
[0.0, self.focal_length, float(self.height)/2],
|
||||
[0.0, 0.0, 1.0]
|
||||
])
|
||||
|
||||
@property
|
||||
def intrinsics_inv(self):
|
||||
# aka 'K_inv' aka view_frame_from_camera_frame
|
||||
return np.linalg.inv(self.intrinsics)
|
||||
|
||||
@dataclass(frozen=True)
|
||||
class _NoneCameraConfig(CameraConfig):
|
||||
width: int = 0
|
||||
height: int = 0
|
||||
focal_length: float = 0
|
||||
|
||||
@dataclass(frozen=True)
|
||||
class DeviceCameraConfig:
|
||||
fcam: CameraConfig
|
||||
dcam: CameraConfig
|
||||
ecam: CameraConfig
|
||||
|
||||
def all_cams(self):
|
||||
for cam in ['fcam', 'dcam', 'ecam']:
|
||||
if not isinstance(getattr(self, cam), _NoneCameraConfig):
|
||||
yield cam, getattr(self, cam)
|
||||
|
||||
_ar_ox_fisheye = CameraConfig(1928, 1208, 567.0) # focal length probably wrong? magnification is not consistent across frame
|
||||
_os_fisheye = CameraConfig(2688 // 2, 1520 // 2, 567.0 / 4 * 3)
|
||||
_ar_ox_config = DeviceCameraConfig(CameraConfig(1928, 1208, 2648.0), _ar_ox_fisheye, _ar_ox_fisheye)
|
||||
_os_config = DeviceCameraConfig(CameraConfig(2688 // 2, 1520 // 2, 1522.0 * 3 / 4), _os_fisheye, _os_fisheye)
|
||||
_neo_config = DeviceCameraConfig(CameraConfig(1164, 874, 910.0), CameraConfig(816, 612, 650.0), _NoneCameraConfig())
|
||||
|
||||
DEVICE_CAMERAS = {
|
||||
# A "device camera" is defined by a device type and sensor
|
||||
|
||||
# sensor type was never set on eon/neo/two
|
||||
("neo", "unknown"): _neo_config,
|
||||
# unknown here is AR0231, field was added with OX03C10 support
|
||||
("tici", "unknown"): _ar_ox_config,
|
||||
|
||||
# before deviceState.deviceType was set, assume tici AR config
|
||||
("unknown", "ar0231"): _ar_ox_config,
|
||||
("unknown", "ox03c10"): _ar_ox_config,
|
||||
|
||||
# simulator (emulates a tici)
|
||||
("pc", "unknown"): _ar_ox_config,
|
||||
}
|
||||
prods = itertools.product(('tici', 'tizi', 'mici'), (('ar0231', _ar_ox_config), ('ox03c10', _ar_ox_config), ('os04c10', _os_config)))
|
||||
DEVICE_CAMERAS.update({(d, c[0]): c[1] for d, c in prods})
|
||||
|
||||
# device/mesh : x->forward, y-> right, z->down
|
||||
# view : x->right, y->down, z->forward
|
||||
device_frame_from_view_frame = np.array([
|
||||
[ 0., 0., 1.],
|
||||
[ 1., 0., 0.],
|
||||
[ 0., 1., 0.]
|
||||
])
|
||||
view_frame_from_device_frame = device_frame_from_view_frame.T
|
||||
|
||||
|
||||
# aka 'extrinsic_matrix'
|
||||
# road : x->forward, y -> left, z->up
|
||||
def get_view_frame_from_road_frame(roll, pitch, yaw, height):
|
||||
device_from_road = orient.rot_from_euler([roll, pitch, yaw]).dot(np.diag([1, -1, -1]))
|
||||
view_from_road = view_frame_from_device_frame.dot(device_from_road)
|
||||
return np.hstack((view_from_road, [[0], [height], [0]]))
|
||||
|
||||
|
||||
|
||||
# aka 'extrinsic_matrix'
|
||||
def get_view_frame_from_calib_frame(roll, pitch, yaw, height):
|
||||
device_from_calib= orient.rot_from_euler([roll, pitch, yaw])
|
||||
view_from_calib = view_frame_from_device_frame.dot(device_from_calib)
|
||||
return np.hstack((view_from_calib, [[0], [height], [0]]))
|
||||
|
||||
|
||||
def vp_from_ke(m):
|
||||
"""
|
||||
Computes the vanishing point from the product of the intrinsic and extrinsic
|
||||
matrices C = KE.
|
||||
|
||||
The vanishing point is defined as lim x->infinity C (x, 0, 0, 1).T
|
||||
"""
|
||||
return (m[0, 0]/m[2, 0], m[1, 0]/m[2, 0])
|
||||
|
||||
|
||||
def roll_from_ke(m):
|
||||
# note: different from calibration.h/RollAnglefromKE: i think that one's just wrong
|
||||
return np.arctan2(-(m[1, 0] - m[1, 1] * m[2, 0] / m[2, 1]),
|
||||
-(m[0, 0] - m[0, 1] * m[2, 0] / m[2, 1]))
|
||||
|
||||
|
||||
def normalize(img_pts, intrinsics):
|
||||
# normalizes image coordinates
|
||||
# accepts single pt or array of pts
|
||||
intrinsics_inv = np.linalg.inv(intrinsics)
|
||||
img_pts = np.array(img_pts)
|
||||
input_shape = img_pts.shape
|
||||
img_pts = np.atleast_2d(img_pts)
|
||||
img_pts = np.hstack((img_pts, np.ones((img_pts.shape[0], 1))))
|
||||
img_pts_normalized = img_pts.dot(intrinsics_inv.T)
|
||||
img_pts_normalized[(img_pts < 0).any(axis=1)] = np.nan
|
||||
return img_pts_normalized[:, :2].reshape(input_shape)
|
||||
|
||||
|
||||
def denormalize(img_pts, intrinsics, width=np.inf, height=np.inf):
|
||||
# denormalizes image coordinates
|
||||
# accepts single pt or array of pts
|
||||
img_pts = np.array(img_pts)
|
||||
input_shape = img_pts.shape
|
||||
img_pts = np.atleast_2d(img_pts)
|
||||
img_pts = np.hstack((img_pts, np.ones((img_pts.shape[0], 1), dtype=img_pts.dtype)))
|
||||
img_pts_denormalized = img_pts.dot(intrinsics.T)
|
||||
if np.isfinite(width):
|
||||
img_pts_denormalized[img_pts_denormalized[:, 0] > width] = np.nan
|
||||
img_pts_denormalized[img_pts_denormalized[:, 0] < 0] = np.nan
|
||||
if np.isfinite(height):
|
||||
img_pts_denormalized[img_pts_denormalized[:, 1] > height] = np.nan
|
||||
img_pts_denormalized[img_pts_denormalized[:, 1] < 0] = np.nan
|
||||
return img_pts_denormalized[:, :2].reshape(input_shape)
|
||||
|
||||
|
||||
def get_calib_from_vp(vp, intrinsics):
|
||||
vp_norm = normalize(vp, intrinsics)
|
||||
yaw_calib = np.arctan(vp_norm[0])
|
||||
pitch_calib = -np.arctan(vp_norm[1]*np.cos(yaw_calib))
|
||||
roll_calib = 0
|
||||
return roll_calib, pitch_calib, yaw_calib
|
||||
|
||||
|
||||
def device_from_ecef(pos_ecef, orientation_ecef, pt_ecef):
|
||||
# device from ecef frame
|
||||
# device frame is x -> forward, y-> right, z -> down
|
||||
# accepts single pt or array of pts
|
||||
input_shape = pt_ecef.shape
|
||||
pt_ecef = np.atleast_2d(pt_ecef)
|
||||
ecef_from_device_rot = orient.rotations_from_quats(orientation_ecef)
|
||||
device_from_ecef_rot = ecef_from_device_rot.T
|
||||
pt_ecef_rel = pt_ecef - pos_ecef
|
||||
pt_device = np.einsum('jk,ik->ij', device_from_ecef_rot, pt_ecef_rel)
|
||||
return pt_device.reshape(input_shape)
|
||||
|
||||
|
||||
def img_from_device(pt_device):
|
||||
# img coordinates from pts in device frame
|
||||
# first transforms to view frame, then to img coords
|
||||
# accepts single pt or array of pts
|
||||
input_shape = pt_device.shape
|
||||
pt_device = np.atleast_2d(pt_device)
|
||||
pt_view = np.einsum('jk,ik->ij', view_frame_from_device_frame, pt_device)
|
||||
|
||||
# This function should never return negative depths
|
||||
pt_view[pt_view[:, 2] < 0] = np.nan
|
||||
|
||||
pt_img = pt_view/pt_view[:, 2:3]
|
||||
return pt_img.reshape(input_shape)[:, :2]
|
||||
|
||||
43
common/transformations/coordinates.hpp
Normal file
43
common/transformations/coordinates.hpp
Normal file
@@ -0,0 +1,43 @@
|
||||
#pragma once
|
||||
|
||||
#include <eigen3/Eigen/Dense>
|
||||
|
||||
#define DEG2RAD(x) ((x) * M_PI / 180.0)
|
||||
#define RAD2DEG(x) ((x) * 180.0 / M_PI)
|
||||
|
||||
struct ECEF {
|
||||
double x, y, z;
|
||||
Eigen::Vector3d to_vector() const {
|
||||
return Eigen::Vector3d(x, y, z);
|
||||
}
|
||||
};
|
||||
|
||||
struct NED {
|
||||
double n, e, d;
|
||||
Eigen::Vector3d to_vector() const {
|
||||
return Eigen::Vector3d(n, e, d);
|
||||
}
|
||||
};
|
||||
|
||||
struct Geodetic {
|
||||
double lat, lon, alt;
|
||||
bool radians=false;
|
||||
};
|
||||
|
||||
ECEF geodetic2ecef(const Geodetic &g);
|
||||
Geodetic ecef2geodetic(const ECEF &e);
|
||||
|
||||
class LocalCoord {
|
||||
public:
|
||||
Eigen::Matrix3d ned2ecef_matrix;
|
||||
Eigen::Matrix3d ecef2ned_matrix;
|
||||
Eigen::Vector3d init_ecef;
|
||||
LocalCoord(const Geodetic &g, const ECEF &e);
|
||||
LocalCoord(const Geodetic &g) : LocalCoord(g, ::geodetic2ecef(g)) {}
|
||||
LocalCoord(const ECEF &e) : LocalCoord(::ecef2geodetic(e), e) {}
|
||||
|
||||
NED ecef2ned(const ECEF &e);
|
||||
ECEF ned2ecef(const NED &n);
|
||||
NED geodetic2ned(const Geodetic &g);
|
||||
Geodetic ned2geodetic(const NED &n);
|
||||
};
|
||||
18
common/transformations/coordinates.py
Normal file
18
common/transformations/coordinates.py
Normal file
@@ -0,0 +1,18 @@
|
||||
from openpilot.common.transformations.orientation import numpy_wrap
|
||||
from openpilot.common.transformations.transformations import (ecef2geodetic_single,
|
||||
geodetic2ecef_single)
|
||||
from openpilot.common.transformations.transformations import LocalCoord as LocalCoord_single
|
||||
|
||||
|
||||
class LocalCoord(LocalCoord_single):
|
||||
ecef2ned = numpy_wrap(LocalCoord_single.ecef2ned_single, (3,), (3,))
|
||||
ned2ecef = numpy_wrap(LocalCoord_single.ned2ecef_single, (3,), (3,))
|
||||
geodetic2ned = numpy_wrap(LocalCoord_single.geodetic2ned_single, (3,), (3,))
|
||||
ned2geodetic = numpy_wrap(LocalCoord_single.ned2geodetic_single, (3,), (3,))
|
||||
|
||||
|
||||
geodetic2ecef = numpy_wrap(geodetic2ecef_single, (3,), (3,))
|
||||
ecef2geodetic = numpy_wrap(ecef2geodetic_single, (3,), (3,))
|
||||
|
||||
geodetic_from_ecef = ecef2geodetic
|
||||
ecef_from_geodetic = geodetic2ecef
|
||||
70
common/transformations/model.py
Normal file
70
common/transformations/model.py
Normal file
@@ -0,0 +1,70 @@
|
||||
import numpy as np
|
||||
|
||||
from openpilot.common.transformations.orientation import rot_from_euler
|
||||
from openpilot.common.transformations.camera import get_view_frame_from_calib_frame, view_frame_from_device_frame, _ar_ox_fisheye
|
||||
|
||||
# segnet
|
||||
SEGNET_SIZE = (512, 384)
|
||||
|
||||
# MED model
|
||||
MEDMODEL_INPUT_SIZE = (512, 256)
|
||||
MEDMODEL_YUV_SIZE = (MEDMODEL_INPUT_SIZE[0], MEDMODEL_INPUT_SIZE[1] * 3 // 2)
|
||||
MEDMODEL_CY = 47.6
|
||||
|
||||
medmodel_fl = 910.0
|
||||
medmodel_intrinsics = np.array([
|
||||
[medmodel_fl, 0.0, 0.5 * MEDMODEL_INPUT_SIZE[0]],
|
||||
[0.0, medmodel_fl, MEDMODEL_CY],
|
||||
[0.0, 0.0, 1.0]])
|
||||
|
||||
|
||||
# BIG model
|
||||
BIGMODEL_INPUT_SIZE = (1024, 512)
|
||||
BIGMODEL_YUV_SIZE = (BIGMODEL_INPUT_SIZE[0], BIGMODEL_INPUT_SIZE[1] * 3 // 2)
|
||||
|
||||
bigmodel_fl = 910.0
|
||||
bigmodel_intrinsics = np.array([
|
||||
[bigmodel_fl, 0.0, 0.5 * BIGMODEL_INPUT_SIZE[0]],
|
||||
[0.0, bigmodel_fl, 256 + MEDMODEL_CY],
|
||||
[0.0, 0.0, 1.0]])
|
||||
|
||||
|
||||
# SBIG model (big model with the size of small model)
|
||||
SBIGMODEL_INPUT_SIZE = (512, 256)
|
||||
SBIGMODEL_YUV_SIZE = (SBIGMODEL_INPUT_SIZE[0], SBIGMODEL_INPUT_SIZE[1] * 3 // 2)
|
||||
|
||||
sbigmodel_fl = 455.0
|
||||
sbigmodel_intrinsics = np.array([
|
||||
[sbigmodel_fl, 0.0, 0.5 * SBIGMODEL_INPUT_SIZE[0]],
|
||||
[0.0, sbigmodel_fl, 0.5 * (256 + MEDMODEL_CY)],
|
||||
[0.0, 0.0, 1.0]])
|
||||
|
||||
DM_INPUT_SIZE = (1440, 960)
|
||||
dmonitoringmodel_fl = _ar_ox_fisheye.focal_length
|
||||
dmonitoringmodel_intrinsics = np.array([
|
||||
[dmonitoringmodel_fl, 0.0, DM_INPUT_SIZE[0]/2],
|
||||
[0.0, dmonitoringmodel_fl, DM_INPUT_SIZE[1]/2 - (_ar_ox_fisheye.height - DM_INPUT_SIZE[1])/2],
|
||||
[0.0, 0.0, 1.0]])
|
||||
|
||||
bigmodel_frame_from_calib_frame = np.dot(bigmodel_intrinsics,
|
||||
get_view_frame_from_calib_frame(0, 0, 0, 0))
|
||||
|
||||
|
||||
sbigmodel_frame_from_calib_frame = np.dot(sbigmodel_intrinsics,
|
||||
get_view_frame_from_calib_frame(0, 0, 0, 0))
|
||||
|
||||
medmodel_frame_from_calib_frame = np.dot(medmodel_intrinsics,
|
||||
get_view_frame_from_calib_frame(0, 0, 0, 0))
|
||||
|
||||
medmodel_frame_from_bigmodel_frame = np.dot(medmodel_intrinsics, np.linalg.inv(bigmodel_intrinsics))
|
||||
|
||||
calib_from_medmodel = np.linalg.inv(medmodel_frame_from_calib_frame[:, :3])
|
||||
calib_from_sbigmodel = np.linalg.inv(sbigmodel_frame_from_calib_frame[:, :3])
|
||||
|
||||
# This function is verified to give similar results to xx.uncommon.utils.transform_img
|
||||
def get_warp_matrix(device_from_calib_euler: np.ndarray, intrinsics: np.ndarray, bigmodel_frame: bool = False) -> np.ndarray:
|
||||
calib_from_model = calib_from_sbigmodel if bigmodel_frame else calib_from_medmodel
|
||||
device_from_calib = rot_from_euler(device_from_calib_euler)
|
||||
camera_from_calib = intrinsics @ view_frame_from_device_frame @ device_from_calib
|
||||
warp_matrix: np.ndarray = camera_from_calib @ calib_from_model
|
||||
return warp_matrix
|
||||
17
common/transformations/orientation.hpp
Normal file
17
common/transformations/orientation.hpp
Normal file
@@ -0,0 +1,17 @@
|
||||
#pragma once
|
||||
#include <eigen3/Eigen/Dense>
|
||||
#include "common/transformations/coordinates.hpp"
|
||||
|
||||
|
||||
Eigen::Quaterniond ensure_unique(const Eigen::Quaterniond &quat);
|
||||
|
||||
Eigen::Quaterniond euler2quat(const Eigen::Vector3d &euler);
|
||||
Eigen::Vector3d quat2euler(const Eigen::Quaterniond &quat);
|
||||
Eigen::Matrix3d quat2rot(const Eigen::Quaterniond &quat);
|
||||
Eigen::Quaterniond rot2quat(const Eigen::Matrix3d &rot);
|
||||
Eigen::Matrix3d euler2rot(const Eigen::Vector3d &euler);
|
||||
Eigen::Vector3d rot2euler(const Eigen::Matrix3d &rot);
|
||||
Eigen::Matrix3d rot_matrix(double roll, double pitch, double yaw);
|
||||
Eigen::Matrix3d rot(const Eigen::Vector3d &axis, double angle);
|
||||
Eigen::Vector3d ecef_euler_from_ned(const ECEF &ecef_init, const Eigen::Vector3d &ned_pose);
|
||||
Eigen::Vector3d ned_euler_from_ecef(const ECEF &ecef_init, const Eigen::Vector3d &ecef_pose);
|
||||
52
common/transformations/orientation.py
Normal file
52
common/transformations/orientation.py
Normal file
@@ -0,0 +1,52 @@
|
||||
import numpy as np
|
||||
from collections.abc import Callable
|
||||
|
||||
from openpilot.common.transformations.transformations import (ecef_euler_from_ned_single,
|
||||
euler2quat_single,
|
||||
euler2rot_single,
|
||||
ned_euler_from_ecef_single,
|
||||
quat2euler_single,
|
||||
quat2rot_single,
|
||||
rot2euler_single,
|
||||
rot2quat_single)
|
||||
|
||||
|
||||
def numpy_wrap(function, input_shape, output_shape) -> Callable[..., np.ndarray]:
|
||||
"""Wrap a function to take either an input or list of inputs and return the correct shape"""
|
||||
def f(*inps):
|
||||
*args, inp = inps
|
||||
inp = np.array(inp)
|
||||
shape = inp.shape
|
||||
|
||||
if len(shape) == len(input_shape):
|
||||
out_shape = output_shape
|
||||
else:
|
||||
out_shape = (shape[0],) + output_shape
|
||||
|
||||
# Add empty dimension if inputs is not a list
|
||||
if len(shape) == len(input_shape):
|
||||
inp.shape = (1, ) + inp.shape
|
||||
|
||||
result = np.asarray([function(*args, i) for i in inp])
|
||||
result.shape = out_shape
|
||||
return result
|
||||
return f
|
||||
|
||||
|
||||
euler2quat = numpy_wrap(euler2quat_single, (3,), (4,))
|
||||
quat2euler = numpy_wrap(quat2euler_single, (4,), (3,))
|
||||
quat2rot = numpy_wrap(quat2rot_single, (4,), (3, 3))
|
||||
rot2quat = numpy_wrap(rot2quat_single, (3, 3), (4,))
|
||||
euler2rot = numpy_wrap(euler2rot_single, (3,), (3, 3))
|
||||
rot2euler = numpy_wrap(rot2euler_single, (3, 3), (3,))
|
||||
ecef_euler_from_ned = numpy_wrap(ecef_euler_from_ned_single, (3,), (3,))
|
||||
ned_euler_from_ecef = numpy_wrap(ned_euler_from_ecef_single, (3,), (3,))
|
||||
|
||||
quats_from_rotations = rot2quat
|
||||
quat_from_rot = rot2quat
|
||||
rotations_from_quats = quat2rot
|
||||
rot_from_quat = quat2rot
|
||||
euler_from_rot = rot2euler
|
||||
euler_from_quat = quat2euler
|
||||
rot_from_euler = euler2rot
|
||||
quat_from_euler = euler2quat
|
||||
0
common/transformations/tests/__init__.py
Normal file
0
common/transformations/tests/__init__.py
Normal file
104
common/transformations/tests/test_coordinates.py
Normal file
104
common/transformations/tests/test_coordinates.py
Normal file
@@ -0,0 +1,104 @@
|
||||
import numpy as np
|
||||
|
||||
import openpilot.common.transformations.coordinates as coord
|
||||
|
||||
geodetic_positions = np.array([[37.7610403, -122.4778699, 115],
|
||||
[27.4840915, -68.5867592, 2380],
|
||||
[32.4916858, -113.652821, -6],
|
||||
[15.1392514, 103.6976037, 24],
|
||||
[24.2302229, 44.2835412, 1650]])
|
||||
|
||||
ecef_positions = np.array([[-2711076.55270557, -4259167.14692758, 3884579.87669935],
|
||||
[ 2068042.69652729, -5273435.40316622, 2927004.89190746],
|
||||
[-2160412.60461669, -4932588.89873832, 3406542.29652851],
|
||||
[-1458247.92550567, 5983060.87496612, 1654984.6099885 ],
|
||||
[ 4167239.10867871, 4064301.90363223, 2602234.6065749 ]])
|
||||
|
||||
ecef_positions_offset = np.array([[-2711004.46961115, -4259099.33540613, 3884605.16002147],
|
||||
[ 2068074.30639499, -5273413.78835412, 2927012.48741131],
|
||||
[-2160344.53748176, -4932586.20092211, 3406636.2962545 ],
|
||||
[-1458211.98517094, 5983151.11161276, 1655077.02698447],
|
||||
[ 4167271.20055269, 4064398.22619263, 2602238.95265847]])
|
||||
|
||||
|
||||
ned_offsets = np.array([[78.722153649976391, 24.396208657446344, 60.343017506838436],
|
||||
[10.699003365155221, 37.319278617604269, 4.1084100025050407],
|
||||
[95.282646251726959, 61.266689955574428, -25.376506058505054],
|
||||
[68.535769283630003, -56.285970011848889, -100.54840137956515],
|
||||
[-33.066609321880179, 46.549821994306861, -84.062540548335591]])
|
||||
|
||||
ecef_init_batch = np.array([2068042.69652729, -5273435.40316622, 2927004.89190746])
|
||||
ecef_positions_offset_batch = np.array([[ 2068089.41454771, -5273434.46829148, 2927074.04783672],
|
||||
[ 2068103.31628647, -5273393.92275431, 2927102.08725987],
|
||||
[ 2068108.49939636, -5273359.27047121, 2927045.07091581],
|
||||
[ 2068075.12395611, -5273381.69432566, 2927041.08207992],
|
||||
[ 2068060.72033399, -5273430.6061505, 2927094.54928305]])
|
||||
|
||||
ned_offsets_batch = np.array([[ 53.88103168, 43.83445935, -46.27488057],
|
||||
[ 93.83378995, 71.57943024, -30.23113187],
|
||||
[ 57.26725796, 89.05602684, 23.02265814],
|
||||
[ 49.71775195, 49.79767572, 17.15351015],
|
||||
[ 78.56272609, 18.53100158, -43.25290759]])
|
||||
|
||||
|
||||
class TestNED:
|
||||
def test_small_distances(self):
|
||||
start_geodetic = np.array([33.8042184, -117.888593, 0.0])
|
||||
local_coord = coord.LocalCoord.from_geodetic(start_geodetic)
|
||||
|
||||
start_ned = local_coord.geodetic2ned(start_geodetic)
|
||||
np.testing.assert_array_equal(start_ned, np.zeros(3,))
|
||||
|
||||
west_geodetic = start_geodetic + [0, -0.0005, 0]
|
||||
west_ned = local_coord.geodetic2ned(west_geodetic)
|
||||
assert np.abs(west_ned[0]) < 1e-3
|
||||
assert west_ned[1] < 0
|
||||
|
||||
southwest_geodetic = start_geodetic + [-0.0005, -0.002, 0]
|
||||
southwest_ned = local_coord.geodetic2ned(southwest_geodetic)
|
||||
assert southwest_ned[0] < 0
|
||||
assert southwest_ned[1] < 0
|
||||
|
||||
def test_ecef_geodetic(self):
|
||||
# testing single
|
||||
np.testing.assert_allclose(ecef_positions[0], coord.geodetic2ecef(geodetic_positions[0]), rtol=1e-9)
|
||||
np.testing.assert_allclose(geodetic_positions[0, :2], coord.ecef2geodetic(ecef_positions[0])[:2], rtol=1e-9)
|
||||
np.testing.assert_allclose(geodetic_positions[0, 2], coord.ecef2geodetic(ecef_positions[0])[2], rtol=1e-9, atol=1e-4)
|
||||
|
||||
np.testing.assert_allclose(geodetic_positions[:, :2], coord.ecef2geodetic(ecef_positions)[:, :2], rtol=1e-9)
|
||||
np.testing.assert_allclose(geodetic_positions[:, 2], coord.ecef2geodetic(ecef_positions)[:, 2], rtol=1e-9, atol=1e-4)
|
||||
np.testing.assert_allclose(ecef_positions, coord.geodetic2ecef(geodetic_positions), rtol=1e-9)
|
||||
|
||||
|
||||
def test_ned(self):
|
||||
for ecef_pos in ecef_positions:
|
||||
converter = coord.LocalCoord.from_ecef(ecef_pos)
|
||||
ecef_pos_moved = ecef_pos + [25, -25, 25]
|
||||
ecef_pos_moved_double_converted = converter.ned2ecef(converter.ecef2ned(ecef_pos_moved))
|
||||
np.testing.assert_allclose(ecef_pos_moved, ecef_pos_moved_double_converted, rtol=1e-9)
|
||||
|
||||
for geo_pos in geodetic_positions:
|
||||
converter = coord.LocalCoord.from_geodetic(geo_pos)
|
||||
geo_pos_moved = geo_pos + np.array([0, 0, 10])
|
||||
geo_pos_double_converted_moved = converter.ned2geodetic(converter.geodetic2ned(geo_pos) + np.array([0, 0, -10]))
|
||||
np.testing.assert_allclose(geo_pos_moved[:2], geo_pos_double_converted_moved[:2], rtol=1e-9, atol=1e-6)
|
||||
np.testing.assert_allclose(geo_pos_moved[2], geo_pos_double_converted_moved[2], rtol=1e-9, atol=1e-4)
|
||||
|
||||
def test_ned_saved_results(self):
|
||||
for i, ecef_pos in enumerate(ecef_positions):
|
||||
converter = coord.LocalCoord.from_ecef(ecef_pos)
|
||||
np.testing.assert_allclose(converter.ned2ecef(ned_offsets[i]),
|
||||
ecef_positions_offset[i],
|
||||
rtol=1e-9, atol=1e-4)
|
||||
np.testing.assert_allclose(converter.ecef2ned(ecef_positions_offset[i]),
|
||||
ned_offsets[i],
|
||||
rtol=1e-9, atol=1e-4)
|
||||
|
||||
def test_ned_batch(self):
|
||||
converter = coord.LocalCoord.from_ecef(ecef_init_batch)
|
||||
np.testing.assert_allclose(converter.ecef2ned(ecef_positions_offset_batch),
|
||||
ned_offsets_batch,
|
||||
rtol=1e-9, atol=1e-7)
|
||||
np.testing.assert_allclose(converter.ned2ecef(ned_offsets_batch),
|
||||
ecef_positions_offset_batch,
|
||||
rtol=1e-9, atol=1e-7)
|
||||
61
common/transformations/tests/test_orientation.py
Normal file
61
common/transformations/tests/test_orientation.py
Normal file
@@ -0,0 +1,61 @@
|
||||
import numpy as np
|
||||
|
||||
from openpilot.common.transformations.orientation import euler2quat, quat2euler, euler2rot, rot2euler, \
|
||||
rot2quat, quat2rot, \
|
||||
ned_euler_from_ecef
|
||||
|
||||
eulers = np.array([[ 1.46520501, 2.78688383, 2.92780854],
|
||||
[ 4.86909526, 3.60618161, 4.30648981],
|
||||
[ 3.72175965, 2.68763705, 5.43895988],
|
||||
[ 5.92306687, 5.69573614, 0.81100357],
|
||||
[ 0.67838374, 5.02402037, 2.47106426]])
|
||||
|
||||
quats = np.array([[ 0.66855182, -0.71500939, 0.19539353, 0.06017818],
|
||||
[ 0.43163717, 0.70013301, 0.28209145, 0.49389021],
|
||||
[ 0.44121991, -0.08252646, 0.34257534, 0.82532207],
|
||||
[ 0.88578382, -0.04515356, -0.32936046, 0.32383617],
|
||||
[ 0.06578165, 0.61282835, 0.07126891, 0.78424163]])
|
||||
|
||||
ecef_positions = np.array([[-2711076.55270557, -4259167.14692758, 3884579.87669935],
|
||||
[ 2068042.69652729, -5273435.40316622, 2927004.89190746],
|
||||
[-2160412.60461669, -4932588.89873832, 3406542.29652851],
|
||||
[-1458247.92550567, 5983060.87496612, 1654984.6099885 ],
|
||||
[ 4167239.10867871, 4064301.90363223, 2602234.6065749 ]])
|
||||
|
||||
ned_eulers = np.array([[ 0.46806039, -0.4881889 , 1.65697808],
|
||||
[-2.14525969, -0.36533066, 0.73813479],
|
||||
[-1.39523364, -0.58540761, -1.77376356],
|
||||
[-1.84220435, 0.61828016, -1.03310421],
|
||||
[ 2.50450101, 0.36304151, 0.33136365]])
|
||||
|
||||
|
||||
class TestOrientation:
|
||||
def test_quat_euler(self):
|
||||
for i, eul in enumerate(eulers):
|
||||
np.testing.assert_allclose(quats[i], euler2quat(eul), rtol=1e-7)
|
||||
np.testing.assert_allclose(quats[i], euler2quat(quat2euler(quats[i])), rtol=1e-6)
|
||||
for i, eul in enumerate(eulers):
|
||||
np.testing.assert_allclose(quats[i], euler2quat(list(eul)), rtol=1e-7)
|
||||
np.testing.assert_allclose(quats[i], euler2quat(quat2euler(list(quats[i]))), rtol=1e-6)
|
||||
np.testing.assert_allclose(quats, euler2quat(eulers), rtol=1e-7)
|
||||
np.testing.assert_allclose(quats, euler2quat(quat2euler(quats)), rtol=1e-6)
|
||||
|
||||
def test_rot_euler(self):
|
||||
for eul in eulers:
|
||||
np.testing.assert_allclose(euler2quat(eul), euler2quat(rot2euler(euler2rot(eul))), rtol=1e-7)
|
||||
for eul in eulers:
|
||||
np.testing.assert_allclose(euler2quat(eul), euler2quat(rot2euler(euler2rot(list(eul)))), rtol=1e-7)
|
||||
np.testing.assert_allclose(euler2quat(eulers), euler2quat(rot2euler(euler2rot(eulers))), rtol=1e-7)
|
||||
|
||||
def test_rot_quat(self):
|
||||
for quat in quats:
|
||||
np.testing.assert_allclose(quat, rot2quat(quat2rot(quat)), rtol=1e-7)
|
||||
for quat in quats:
|
||||
np.testing.assert_allclose(quat, rot2quat(quat2rot(list(quat))), rtol=1e-7)
|
||||
np.testing.assert_allclose(quats, rot2quat(quat2rot(quats)), rtol=1e-7)
|
||||
|
||||
def test_euler_ned(self):
|
||||
for i in range(len(eulers)):
|
||||
np.testing.assert_allclose(ned_eulers[i], ned_euler_from_ecef(ecef_positions[i], eulers[i]), rtol=1e-7)
|
||||
#np.testing.assert_allclose(eulers[i], ecef_euler_from_ned(ecef_positions[i], ned_eulers[i]), rtol=1e-7)
|
||||
# np.testing.assert_allclose(ned_eulers, ned_euler_from_ecef(ecef_positions, eulers), rtol=1e-7)
|
||||
72
common/transformations/transformations.pxd
Normal file
72
common/transformations/transformations.pxd
Normal file
@@ -0,0 +1,72 @@
|
||||
# cython: language_level=3
|
||||
from libcpp cimport bool
|
||||
|
||||
cdef extern from "orientation.cc":
|
||||
pass
|
||||
|
||||
cdef extern from "orientation.hpp":
|
||||
cdef cppclass Quaternion "Eigen::Quaterniond":
|
||||
Quaternion()
|
||||
Quaternion(double, double, double, double)
|
||||
double w()
|
||||
double x()
|
||||
double y()
|
||||
double z()
|
||||
|
||||
cdef cppclass Vector3 "Eigen::Vector3d":
|
||||
Vector3()
|
||||
Vector3(double, double, double)
|
||||
double operator()(int)
|
||||
|
||||
cdef cppclass Matrix3 "Eigen::Matrix3d":
|
||||
Matrix3()
|
||||
Matrix3(double*)
|
||||
|
||||
double operator()(int, int)
|
||||
|
||||
Quaternion euler2quat(const Vector3 &)
|
||||
Vector3 quat2euler(const Quaternion &)
|
||||
Matrix3 quat2rot(const Quaternion &)
|
||||
Quaternion rot2quat(const Matrix3 &)
|
||||
Vector3 rot2euler(const Matrix3 &)
|
||||
Matrix3 euler2rot(const Vector3 &)
|
||||
Matrix3 rot_matrix(double, double, double)
|
||||
Vector3 ecef_euler_from_ned(const ECEF &, const Vector3 &)
|
||||
Vector3 ned_euler_from_ecef(const ECEF &, const Vector3 &)
|
||||
|
||||
|
||||
cdef extern from "coordinates.cc":
|
||||
cdef struct ECEF:
|
||||
double x
|
||||
double y
|
||||
double z
|
||||
|
||||
cdef struct NED:
|
||||
double n
|
||||
double e
|
||||
double d
|
||||
|
||||
cdef struct Geodetic:
|
||||
double lat
|
||||
double lon
|
||||
double alt
|
||||
bool radians
|
||||
|
||||
ECEF geodetic2ecef(const Geodetic &)
|
||||
Geodetic ecef2geodetic(const ECEF &)
|
||||
|
||||
cdef cppclass LocalCoord_c "LocalCoord":
|
||||
Matrix3 ned2ecef_matrix
|
||||
Matrix3 ecef2ned_matrix
|
||||
|
||||
LocalCoord_c(const Geodetic &, const ECEF &)
|
||||
LocalCoord_c(const Geodetic &)
|
||||
LocalCoord_c(const ECEF &)
|
||||
|
||||
NED ecef2ned(const ECEF &)
|
||||
ECEF ned2ecef(const NED &)
|
||||
NED geodetic2ned(const Geodetic &)
|
||||
Geodetic ned2geodetic(const NED &)
|
||||
|
||||
cdef extern from "coordinates.hpp":
|
||||
pass
|
||||
173
common/transformations/transformations.pyx
Normal file
173
common/transformations/transformations.pyx
Normal file
@@ -0,0 +1,173 @@
|
||||
# distutils: language = c++
|
||||
# cython: language_level = 3
|
||||
from openpilot.common.transformations.transformations cimport Matrix3, Vector3, Quaternion
|
||||
from openpilot.common.transformations.transformations cimport ECEF, NED, Geodetic
|
||||
|
||||
from openpilot.common.transformations.transformations cimport euler2quat as euler2quat_c
|
||||
from openpilot.common.transformations.transformations cimport quat2euler as quat2euler_c
|
||||
from openpilot.common.transformations.transformations cimport quat2rot as quat2rot_c
|
||||
from openpilot.common.transformations.transformations cimport rot2quat as rot2quat_c
|
||||
from openpilot.common.transformations.transformations cimport euler2rot as euler2rot_c
|
||||
from openpilot.common.transformations.transformations cimport rot2euler as rot2euler_c
|
||||
from openpilot.common.transformations.transformations cimport rot_matrix as rot_matrix_c
|
||||
from openpilot.common.transformations.transformations cimport ecef_euler_from_ned as ecef_euler_from_ned_c
|
||||
from openpilot.common.transformations.transformations cimport ned_euler_from_ecef as ned_euler_from_ecef_c
|
||||
from openpilot.common.transformations.transformations cimport geodetic2ecef as geodetic2ecef_c
|
||||
from openpilot.common.transformations.transformations cimport ecef2geodetic as ecef2geodetic_c
|
||||
from openpilot.common.transformations.transformations cimport LocalCoord_c
|
||||
|
||||
|
||||
import numpy as np
|
||||
cimport numpy as np
|
||||
|
||||
cdef np.ndarray[double, ndim=2] matrix2numpy(Matrix3 m):
|
||||
return np.array([
|
||||
[m(0, 0), m(0, 1), m(0, 2)],
|
||||
[m(1, 0), m(1, 1), m(1, 2)],
|
||||
[m(2, 0), m(2, 1), m(2, 2)],
|
||||
])
|
||||
|
||||
cdef Matrix3 numpy2matrix(np.ndarray[double, ndim=2, mode="fortran"] m):
|
||||
assert m.shape[0] == 3
|
||||
assert m.shape[1] == 3
|
||||
return Matrix3(<double*>m.data)
|
||||
|
||||
cdef ECEF list2ecef(ecef):
|
||||
cdef ECEF e
|
||||
e.x = ecef[0]
|
||||
e.y = ecef[1]
|
||||
e.z = ecef[2]
|
||||
return e
|
||||
|
||||
cdef NED list2ned(ned):
|
||||
cdef NED n
|
||||
n.n = ned[0]
|
||||
n.e = ned[1]
|
||||
n.d = ned[2]
|
||||
return n
|
||||
|
||||
cdef Geodetic list2geodetic(geodetic):
|
||||
cdef Geodetic g
|
||||
g.lat = geodetic[0]
|
||||
g.lon = geodetic[1]
|
||||
g.alt = geodetic[2]
|
||||
return g
|
||||
|
||||
def euler2quat_single(euler):
|
||||
cdef Vector3 e = Vector3(euler[0], euler[1], euler[2])
|
||||
cdef Quaternion q = euler2quat_c(e)
|
||||
return [q.w(), q.x(), q.y(), q.z()]
|
||||
|
||||
def quat2euler_single(quat):
|
||||
cdef Quaternion q = Quaternion(quat[0], quat[1], quat[2], quat[3])
|
||||
cdef Vector3 e = quat2euler_c(q)
|
||||
return [e(0), e(1), e(2)]
|
||||
|
||||
def quat2rot_single(quat):
|
||||
cdef Quaternion q = Quaternion(quat[0], quat[1], quat[2], quat[3])
|
||||
cdef Matrix3 r = quat2rot_c(q)
|
||||
return matrix2numpy(r)
|
||||
|
||||
def rot2quat_single(rot):
|
||||
cdef Matrix3 r = numpy2matrix(np.asfortranarray(rot, dtype=np.double))
|
||||
cdef Quaternion q = rot2quat_c(r)
|
||||
return [q.w(), q.x(), q.y(), q.z()]
|
||||
|
||||
def euler2rot_single(euler):
|
||||
cdef Vector3 e = Vector3(euler[0], euler[1], euler[2])
|
||||
cdef Matrix3 r = euler2rot_c(e)
|
||||
return matrix2numpy(r)
|
||||
|
||||
def rot2euler_single(rot):
|
||||
cdef Matrix3 r = numpy2matrix(np.asfortranarray(rot, dtype=np.double))
|
||||
cdef Vector3 e = rot2euler_c(r)
|
||||
return [e(0), e(1), e(2)]
|
||||
|
||||
def rot_matrix(roll, pitch, yaw):
|
||||
return matrix2numpy(rot_matrix_c(roll, pitch, yaw))
|
||||
|
||||
def ecef_euler_from_ned_single(ecef_init, ned_pose):
|
||||
cdef ECEF init = list2ecef(ecef_init)
|
||||
cdef Vector3 pose = Vector3(ned_pose[0], ned_pose[1], ned_pose[2])
|
||||
|
||||
cdef Vector3 e = ecef_euler_from_ned_c(init, pose)
|
||||
return [e(0), e(1), e(2)]
|
||||
|
||||
def ned_euler_from_ecef_single(ecef_init, ecef_pose):
|
||||
cdef ECEF init = list2ecef(ecef_init)
|
||||
cdef Vector3 pose = Vector3(ecef_pose[0], ecef_pose[1], ecef_pose[2])
|
||||
|
||||
cdef Vector3 e = ned_euler_from_ecef_c(init, pose)
|
||||
return [e(0), e(1), e(2)]
|
||||
|
||||
def geodetic2ecef_single(geodetic):
|
||||
cdef Geodetic g = list2geodetic(geodetic)
|
||||
cdef ECEF e = geodetic2ecef_c(g)
|
||||
return [e.x, e.y, e.z]
|
||||
|
||||
def ecef2geodetic_single(ecef):
|
||||
cdef ECEF e = list2ecef(ecef)
|
||||
cdef Geodetic g = ecef2geodetic_c(e)
|
||||
return [g.lat, g.lon, g.alt]
|
||||
|
||||
|
||||
cdef class LocalCoord:
|
||||
cdef LocalCoord_c * lc
|
||||
|
||||
def __init__(self, geodetic=None, ecef=None):
|
||||
assert (geodetic is not None) or (ecef is not None)
|
||||
if geodetic is not None:
|
||||
self.lc = new LocalCoord_c(list2geodetic(geodetic))
|
||||
elif ecef is not None:
|
||||
self.lc = new LocalCoord_c(list2ecef(ecef))
|
||||
|
||||
@property
|
||||
def ned2ecef_matrix(self):
|
||||
return matrix2numpy(self.lc.ned2ecef_matrix)
|
||||
|
||||
@property
|
||||
def ecef2ned_matrix(self):
|
||||
return matrix2numpy(self.lc.ecef2ned_matrix)
|
||||
|
||||
@property
|
||||
def ned_from_ecef_matrix(self):
|
||||
return self.ecef2ned_matrix
|
||||
|
||||
@property
|
||||
def ecef_from_ned_matrix(self):
|
||||
return self.ned2ecef_matrix
|
||||
|
||||
@classmethod
|
||||
def from_geodetic(cls, geodetic):
|
||||
return cls(geodetic=geodetic)
|
||||
|
||||
@classmethod
|
||||
def from_ecef(cls, ecef):
|
||||
return cls(ecef=ecef)
|
||||
|
||||
def ecef2ned_single(self, ecef):
|
||||
assert self.lc
|
||||
cdef ECEF e = list2ecef(ecef)
|
||||
cdef NED n = self.lc.ecef2ned(e)
|
||||
return [n.n, n.e, n.d]
|
||||
|
||||
def ned2ecef_single(self, ned):
|
||||
assert self.lc
|
||||
cdef NED n = list2ned(ned)
|
||||
cdef ECEF e = self.lc.ned2ecef(n)
|
||||
return [e.x, e.y, e.z]
|
||||
|
||||
def geodetic2ned_single(self, geodetic):
|
||||
assert self.lc
|
||||
cdef Geodetic g = list2geodetic(geodetic)
|
||||
cdef NED n = self.lc.geodetic2ned(g)
|
||||
return [n.n, n.e, n.d]
|
||||
|
||||
def ned2geodetic_single(self, ned):
|
||||
assert self.lc
|
||||
cdef NED n = list2ned(ned)
|
||||
cdef Geodetic g = self.lc.ned2geodetic(n)
|
||||
return [g.lat, g.lon, g.alt]
|
||||
|
||||
def __dealloc__(self):
|
||||
del self.lc
|
||||
BIN
common/transformations/transformations.so
Executable file
BIN
common/transformations/transformations.so
Executable file
Binary file not shown.
Reference in New Issue
Block a user